 |
My primary research goals are to develop and apply methods to characterize and compare the structure of compounds using NMR spectroscopy. NMR is applicable to a wide range of materials, including gases, solutions, crystalline solids, amorphous materials and surfaces, and a large number of nuclear isotopes are available as spectroscopic probes, including 1H, 13C and 31P, and over 80 other NMR-accessible nuclei. The NMR experiment is very sensitive to subtle changes in the electronic or geometric environment around each nucleus, making NMR an ideal structural probe. Several specific areas of research are being actively pursued.
We are applying a combination of solid-state and solution NMR techniques in concert to permit in situ analyses of supported species. Application of special techniques such as magic angle spinning as well as refinement of some standard NMR experiments to the unique aspects of swollen resin gels have allowed us to obtain substantial improvements in suppression of background polymer signals as well as improved selectivity in multidimensional correlation techniques. One of the most successful areas to which high-resolution NMR has been applied is the determination of polypeptide structures in solution by multidimensional techniques such as TOCSY, NOESY and HMQC, thus our work using HRMAS NMR of solid-supported peptides is an important extension of the determination of peptide structures to include samples that are still attached to the polymer support. As one example of its utility, NMR spectroscopy of these samples is of interest because many solid-supported synthetic strategies encounter reaction difficulties, such as the so-called "difficult couplings" in peptide synthesis, where reaction efficiency drops precipitously at various stages of the solid-phase coupling reactions. It is believed that this is due to the formation of secondary structure by the growing peptide chain, although no direct evidence exists. Even if secondary structure is the culprit, it is unclear whether the coupling inefficiency results from direct blockage of the reaction site due to peptide folding, or from solvent inaccessibility due to an increase in polymer rigidity and concomitant "deswelling" of the resin. Clearly an in situ structure determination technique is needed, and the ability of NMR to contribute to this area is significant, with its ready application to solution, solid and semi-solids like membranes extensively documented in the past. We initiated studies on several known problematic peptide sequences, most notably acyl carrier peptide (ACP) on Wang resin, which is an established sequence used in testing for difficult couplings. We have found that pulse sequences such as TOCSY and ROESY provide particularly "clean" spectra of epitopes, clear of interfering correlations from background polymer signals. We have furthered this approach by using a TOCSY mixing sequence, DIPSI-2, in concert with a normal one-dimensional pulse-acquire sequence to remove, or at least substantially attenuate, the residual proton signal that arises from the polymer. Recently we have extended our spectroscopic tools by developing inverse-HMQC sequences tailored to the conditions encountered with these heterogeneous spinning samples. It is becoming increasingly clear through this work that the Fmoc protecting group is responsible for many of the interactions that appear spectroscopically when difficult couplings arise.
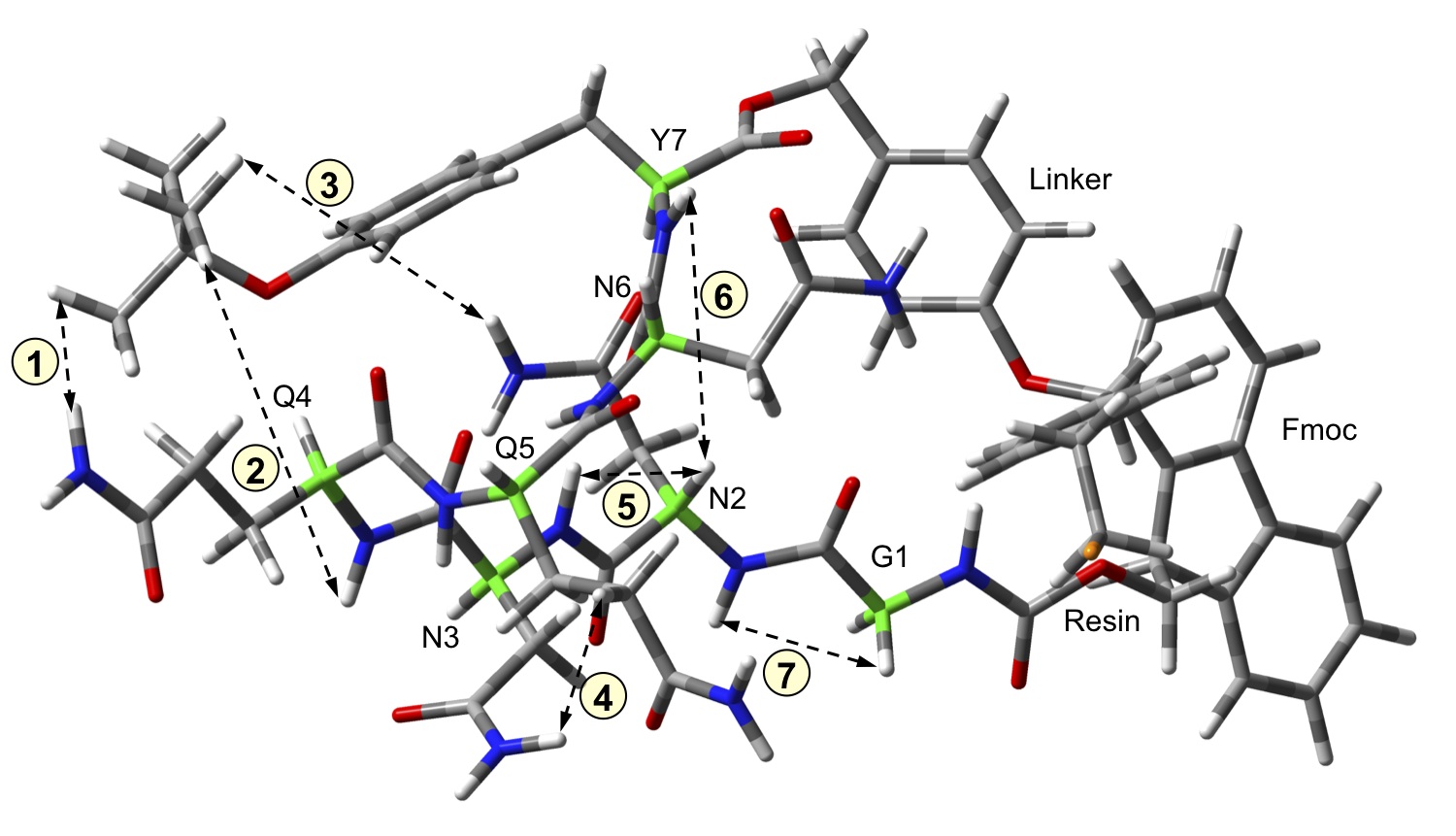
One novel approach we have recently developed is the use of the polymer support as an isolating medium for the individual peptide chains. A short sequence from the yeast prion protein Sup35 (Gly-Asn-Asn-Gln-Gln-Asn-Tyr) has been shown to produce fibrils typical of amyloidosis from solution; however, this very feature makes investigation of the isolated peptide chains impossible to determine in traditional solution experiments. By tethering this peptide chain to a polymer support, we are able to prevent aggregation from occurring, and have provided the first experimental evidence of at least one monomeric conformation.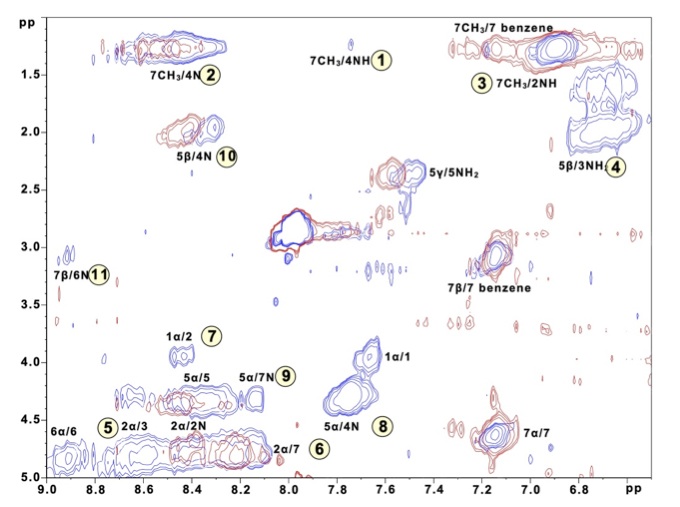 |
 |
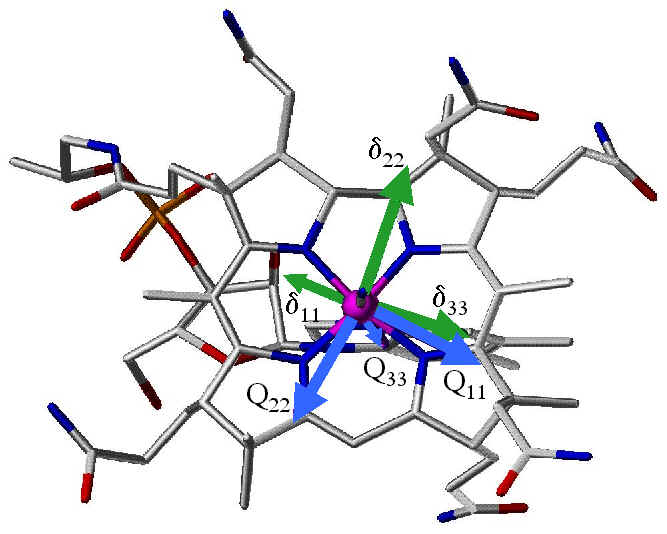
Top View |
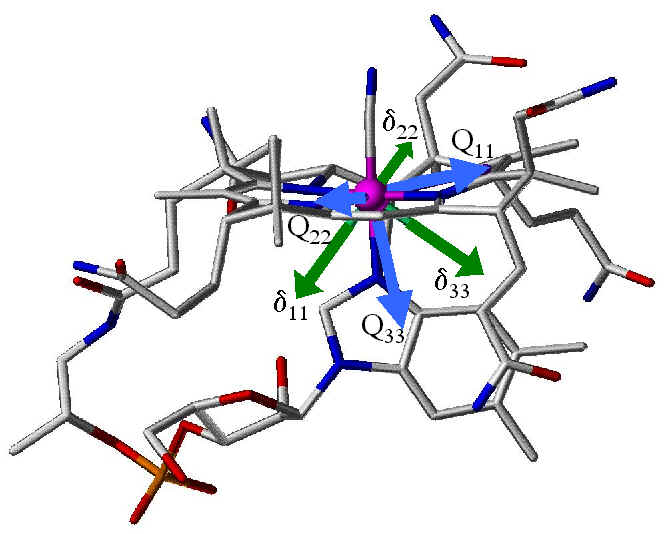
Side View |
|---|---|
 |
 |
 |